Wasserstein distance user manual¶
Definition¶
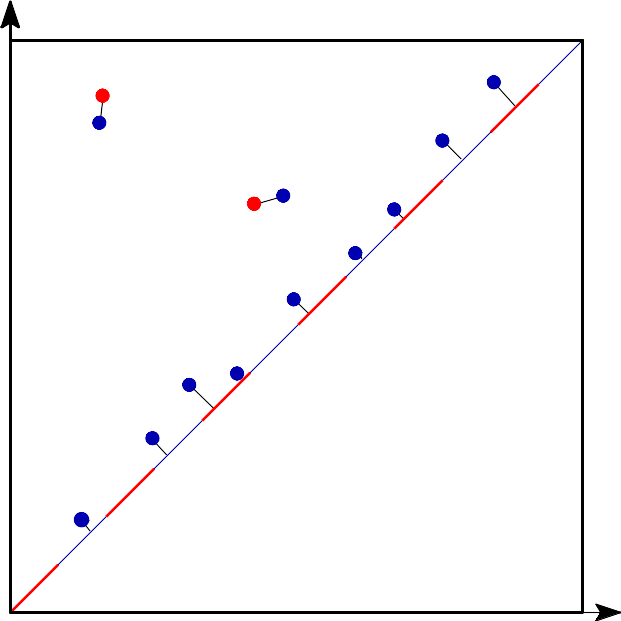
Wasserstein distance is the q-th root of the sum of the edge lengths to the power q.¶ |
The q-Wasserstein distance measures the similarity between two persistence diagrams. It’s the minimum value c that can be achieved by a perfect matching between the points of the two diagrams (+ all diagonal points), where the value of a matching is defined as the q-th root of the sum of all edge lengths to the power q. Edge lengths are measured in norm p, for \(1 \leq p \leq \infty\). |
|
This implementation is based on ideas from “Large Scale Computation of Means and Cluster for Persistence Diagrams via Optimal Transport”.
Function¶
-
gudhi.wasserstein.wasserstein_distance(X, Y, order=2.0, internal_p=2.0)[source]¶ - Parameters
X¶ – (n x 2) numpy.array encoding the (finite points of the) first diagram. Must not contain essential points (i.e. with infinite coordinate).
Y¶ – (m x 2) numpy.array encoding the second diagram.
internal_p¶ – Ground metric on the (upper-half) plane (i.e. norm l_p in R^2); Default value is 2 (euclidean norm).
order¶ – exponent for Wasserstein; Default value is 2.
- Returns
the Wasserstein distance of order q (1 <= q < infinity) between persistence diagrams with respect to the internal_p-norm as ground metric.
- Return type
float
Basic example¶
This example computes the 1-Wasserstein distance from 2 persistence diagrams with euclidean ground metric. Note that persistence diagrams must be submitted as (n x 2) numpy arrays and must not contain inf values.
import gudhi.wasserstein
import numpy as np
diag1 = np.array([[2.7, 3.7],[9.6, 14.],[34.2, 34.974]])
diag2 = np.array([[2.8, 4.45],[9.5, 14.1]])
message = "Wasserstein distance value = " + '%.2f' % gudhi.wasserstein.wasserstein_distance(diag1, diag2, order=1., internal_p=2.)
print(message)
The output is:
Wasserstein distance value = 1.45