- Author
- Vincent Rouvreau
Definition
Alpha_complex is a simplicial complex constructed from the finite cells of a Delaunay Triangulation.
The filtration value of each simplex is computed as the square of the circumradius of the simplex if the circumsphere is empty (the simplex is then said to be Gabriel), and as the minimum of the filtration values of the codimension 1 cofaces that make it not Gabriel otherwise.
All simplices that have a filtration value strictly greater than a given alpha squared value are not inserted into the complex.
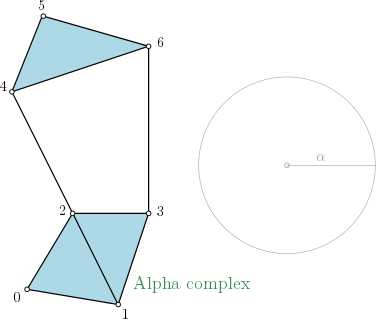
Alpha-complex representation
Alpha_complex is constructing a Delaunay Triangulation [30] from CGAL (the Computational Geometry Algorithms Library [39]) and is able to create a SimplicialComplexForAlpha.
The complex is a template class requiring an Epick_d dD Geometry Kernel [37] from CGAL as template parameter.
Example from points
This example builds the Delaunay triangulation from the given points in a 2D static kernel, and creates a Simplex_tree with it.
Then, it is asked to display information about the simplicial complex.
#include <gudhi/Alpha_complex.h>
#include <gudhi/Simplex_tree.h>
#include <CGAL/Epick_d.h>
#include <iostream>
#include <vector>
using Kernel = CGAL::Epick_d< CGAL::Dimension_tag<2> >;
using Point = Kernel::Point_d;
using Vector_of_points = std::vector<Point>;
int main() {
Vector_of_points points;
points.push_back(Point(1.0, 1.0));
points.push_back(Point(7.0, 0.0));
points.push_back(Point(4.0, 6.0));
points.push_back(Point(9.0, 6.0));
points.push_back(Point(0.0, 14.0));
points.push_back(Point(2.0, 19.0));
points.push_back(Point(9.0, 17.0));
if (alpha_complex_from_points.create_complex(simplex)) {
std::cout <<
"Alpha complex is of dimension " << simplex.
dimension() <<
std::cout << "Iterator on alpha complex simplices in the filtration order, with [filtration value]:" << std::endl;
std::cout << " ( ";
std::cout << vertex << " ";
}
std::cout <<
") -> " <<
"[" << simplex.
filtration(f_simplex) <<
"] ";
std::cout << std::endl;
}
}
return 0;
}
When launching:
$> ./Alpha_complex_example_from_points
the program output is:
Alpha complex is of dimension 2 - 25 simplices - 7 vertices.
Iterator on alpha complex simplices in the filtration order, with [filtration value]:
( 0 ) -> [0]
( 1 ) -> [0]
( 2 ) -> [0]
( 3 ) -> [0]
( 4 ) -> [0]
( 5 ) -> [0]
( 6 ) -> [0]
( 3 2 ) -> [6.25]
( 5 4 ) -> [7.25]
( 2 0 ) -> [8.5]
( 1 0 ) -> [9.25]
( 3 1 ) -> [10]
( 2 1 ) -> [11.25]
( 3 2 1 ) -> [12.5]
( 2 1 0 ) -> [12.9959]
( 6 5 ) -> [13.25]
( 4 2 ) -> [20]
( 6 4 ) -> [22.7367]
( 6 5 4 ) -> [22.7367]
( 6 3 ) -> [30.25]
( 6 2 ) -> [36.5]
( 6 3 2 ) -> [36.5]
( 6 4 2 ) -> [37.2449]
( 4 0 ) -> [59.7107]
( 4 2 0 ) -> [59.7107]
Create complex algorithm
Data structure
In order to create the simplicial complex, first, it is built from the cells of the Delaunay Triangulation. The filtration values are set to NaN, which stands for unknown value.
In example, :
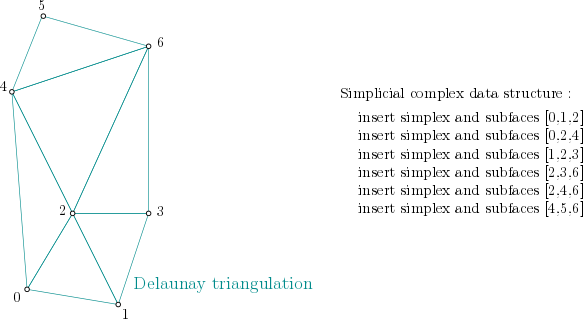
Simplicial complex structure construction example
Filtration value computation algorithm
\( \textbf{for } \text{i : dimension } \rightarrow 0 \textbf{ do}\\ \quad \textbf{for all } \sigma \text{ of dimension i}\\ \quad\quad \textbf{if } \text{filtration(} \sigma ) \text{ is NaN} \textbf{ then}\\ \quad\quad\quad \text{filtration(} \sigma ) = \alpha^2( \sigma )\\ \quad\quad \textbf{end if}\\ \quad\quad \textbf{for all } \tau \text{ face of } \sigma \textbf{ do}\quad\quad \textit{// propagate alpha filtration value}\\ \quad\quad\quad \textbf{if } \text{filtration(} \tau ) \text{ is not NaN} \textbf{ then}\\ \quad\quad\quad\quad \text{filtration(} \tau \text{) = min( filtration(} \tau \text{), filtration(} \sigma \text{) )}\\ \quad\quad\quad \textbf{else}\\ \quad\quad\quad\quad \textbf{if } \tau \text{ is not Gabriel for } \sigma \textbf{ then}\\ \quad\quad\quad\quad\quad \text{filtration(} \tau \text{) = filtration(} \sigma \text{)}\\ \quad\quad\quad\quad \textbf{end if}\\ \quad\quad\quad \textbf{end if}\\ \quad\quad \textbf{end for}\\ \quad \textbf{end for}\\ \textbf{end for}\\ \text{make_filtration_non_decreasing()}\\ \text{prune_above_filtration()}\\ \)
Dimension 2
From the example above, it means the algorithm looks into each triangle ([0,1,2], [0,2,4], [1,2,3], ...), computes the filtration value of the triangle, and then propagates the filtration value as described here :
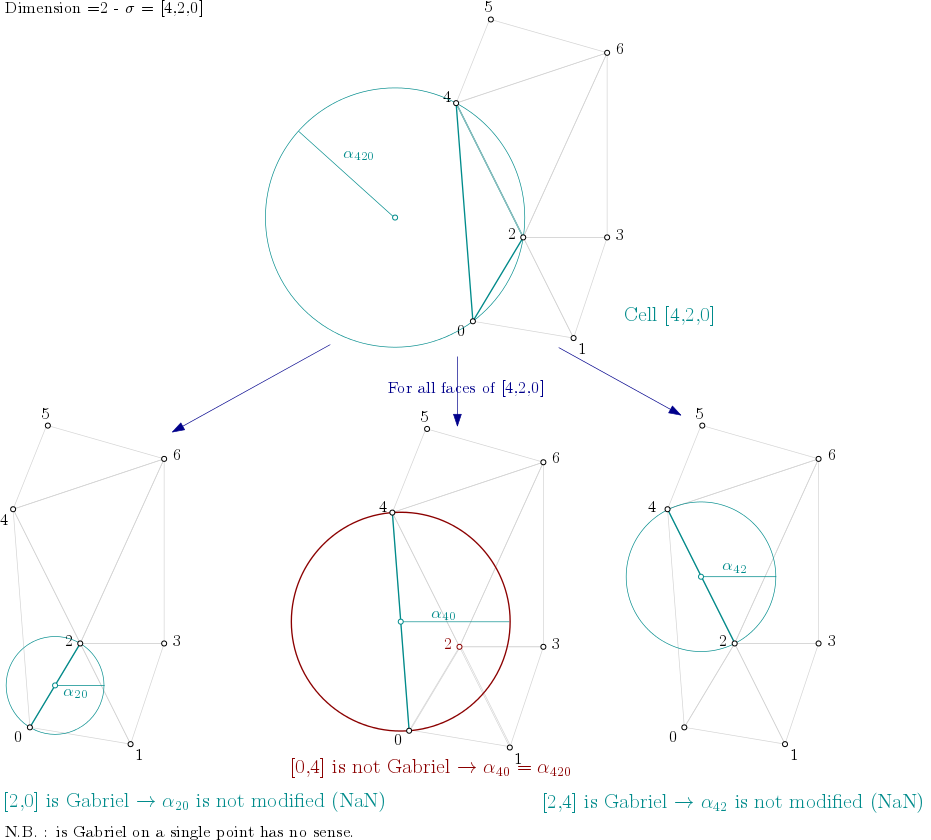
Filtration value propagation example
Dimension 1
Then, the algorithm looks into each edge ([0,1], [0,2], [1,2], ...), computes the filtration value of the edge (in this case, propagation will have no effect).
Dimension 0
Finally, the algorithm looks into each vertex ([0], [1], [2], [3], [4], [5] and [6]) and sets the filtration value (0 in case of a vertex - propagation will have no effect).
Non decreasing filtration values
As the squared radii computed by CGAL are an approximation, it might happen that these alpha squared values do not quite define a proper filtration (i.e. non-decreasing with respect to inclusion). We fix that up by calling SimplicialComplexForAlpha::make_filtration_non_decreasing().
Prune above given filtration value
The simplex tree is pruned from the given maximum alpha squared value (cf. SimplicialComplexForAlpha::prune_above_filtration()). In the following example, the value is given by the user as argument of the program.
Example from OFF file
This example builds the Delaunay triangulation in a dynamic kernel, and initializes the alpha complex with it.
Then, it is asked to display information about the alpha complex.
#include <gudhi/Alpha_complex.h>
#include <gudhi/Simplex_tree.h>
#include <CGAL/Epick_d.h>
#include <iostream>
#include <string>
void usage(int nbArgs, char * const progName) {
std::cerr << "Error: Number of arguments (" << nbArgs << ") is not correct\n";
std::cerr << "Usage: " << progName << " filename.off alpha_square_max_value [ouput_file.txt]\n";
std::cerr << " i.e.: " << progName << " ../../data/points/alphacomplexdoc.off 60.0\n";
exit(-1);
}
int main(int argc, char **argv) {
if ((argc != 3) && (argc != 4)) usage(argc, (argv[0] - 1));
std::string off_file_name {argv[1]};
double alpha_square_max_value {atof(argv[2])};
using Kernel = CGAL::Epick_d< CGAL::Dynamic_dimension_tag >;
std::streambuf* streambufffer;
std::ofstream ouput_file_stream;
if (argc == 4) {
ouput_file_stream.open(std::string(argv[3]));
streambufffer = ouput_file_stream.rdbuf();
} else {
streambufffer = std::cout.rdbuf();
}
if (alpha_complex_from_file.create_complex(simplex, alpha_square_max_value)) {
std::ostream output_stream(streambufffer);
output_stream <<
"Alpha complex is of dimension " << simplex.
dimension() <<
output_stream << "Iterator on alpha complex simplices in the filtration order, with [filtration value]:" <<
std::endl;
output_stream << " ( ";
output_stream << vertex << " ";
}
output_stream <<
") -> " <<
"[" << simplex.
filtration(f_simplex) <<
"] ";
output_stream << std::endl;
}
}
ouput_file_stream.close();
return 0;
}
When launching:
$> ./Alpha_complex_example_from_off ../../data/points/alphacomplexdoc.off 32.0
the program output is:
Alpha complex is of dimension 2 - 20 simplices - 7 vertices.
Iterator on alpha complex simplices in the filtration order, with [filtration value]:
( 0 ) -> [0]
( 1 ) -> [0]
( 2 ) -> [0]
( 3 ) -> [0]
( 4 ) -> [0]
( 5 ) -> [0]
( 6 ) -> [0]
( 3 2 ) -> [6.25]
( 5 4 ) -> [7.25]
( 2 0 ) -> [8.5]
( 1 0 ) -> [9.25]
( 3 1 ) -> [10]
( 2 1 ) -> [11.25]
( 3 2 1 ) -> [12.5]
( 2 1 0 ) -> [12.9959]
( 6 5 ) -> [13.25]
( 4 2 ) -> [20]
( 6 4 ) -> [22.7367]
( 6 5 4 ) -> [22.7367]
( 6 3 ) -> [30.25]
3d specific example
A specific module for Alpha complex is available in 3d (cf. Alpha_complex_3d) and allows to construct standard, weighted, periodic or weighted and periodic versions of alpha complexes. Alpha values computation can be Gudhi::alpha_complex::complexity::FAST, Gudhi::alpha_complex::complexity::SAFE (default value) or Gudhi::alpha_complex::complexity::EXACT.
This example builds the CGAL 3d weighted alpha shapes from a small molecule, and initializes the alpha complex with it. This example is taken from CGAL 3d weighted alpha shapes.
Then, it is asked to display information about the alpha complex.
#include <gudhi/Alpha_complex_3d.h>
#include <gudhi/Simplex_tree.h>
#include <iostream>
#include <string>
#include <vector>
#include <limits>
using Weighted_alpha_complex_3d =
using Point = Weighted_alpha_complex_3d::Point_3;
using Weighted_point = Weighted_alpha_complex_3d::Weighted_point_3;
int main(int argc, char **argv) {
std::vector<Weighted_point> weighted_points;
weighted_points.push_back(Weighted_point(Point(1, -1, -1), 4.));
weighted_points.push_back(Weighted_point(Point(-1, 1, -1), 4.));
weighted_points.push_back(Weighted_point(Point(-1, -1, 1), 4.));
weighted_points.push_back(Weighted_point(Point(1, 1, 1), 4.));
weighted_points.push_back(Weighted_point(Point(2, 2, 2), 1.));
Weighted_alpha_complex_3d alpha_complex_from_points(weighted_points);
if (alpha_complex_from_points.create_complex(simplex)) {
<<
" simplices - " << simplex.
num_vertices() <<
" vertices." << std::endl;
std::cout << "Iterator on alpha complex simplices in the filtration order, with [filtration value]:" << std::endl;
std::cout << " ( ";
std::cout << vertex << " ";
}
std::cout << ") -> "
std::cout << std::endl;
}
}
return 0;
}
When launching:
$> ./Alpha_complex_example_weighted_3d_from_points
the program output is:
Alpha complex is of dimension 3 - 29 simplices - 5 vertices.
Iterator on alpha complex simplices in the filtration order, with [filtration value]:
( 0 ) -> [-4]
( 1 ) -> [-4]
( 2 ) -> [-4]
( 3 ) -> [-4]
( 1 0 ) -> [-2]
( 2 0 ) -> [-2]
( 2 1 ) -> [-2]
( 3 0 ) -> [-2]
( 3 1 ) -> [-2]
( 3 2 ) -> [-2]
( 2 1 0 ) -> [-1.33333]
( 3 1 0 ) -> [-1.33333]
( 3 2 0 ) -> [-1.33333]
( 3 2 1 ) -> [-1.33333]
( 3 2 1 0 ) -> [-1]
( 4 ) -> [-1]
( 4 2 ) -> [-1]
( 4 0 ) -> [23]
( 4 1 ) -> [23]
( 4 2 0 ) -> [23]
( 4 2 1 ) -> [23]
( 4 3 ) -> [23]
( 4 3 2 ) -> [23]
( 4 1 0 ) -> [95]
( 4 2 1 0 ) -> [95]
( 4 3 0 ) -> [95]
( 4 3 1 ) -> [95]
( 4 3 2 0 ) -> [95]
( 4 3 2 1 ) -> [95]
◆ complexity


